Mobile DNA: the clay that genomes are (mostly) made of
Our research focuses on mobile DNA, a term that encompasses diverse genetic elements that were at least once able to integrate and often propagate within the genome. In eukaryotes, these include transposable elements (aka transposons) and endogenous viruses.
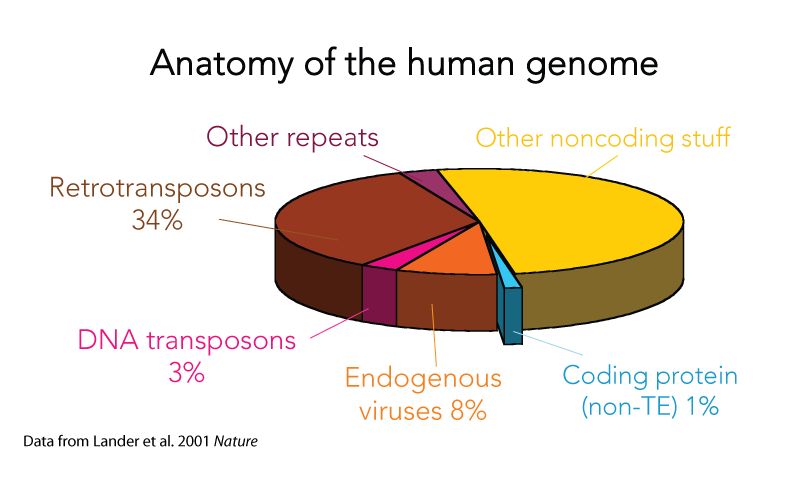
Mobile DNA is found in nearly all organisms and often account for the bulk of the genetic material of a species. For example, the initial characterization of the human genome revealed that transposons and their remnants make up at least 40% of the genome and endogenous retroviruses account for another 8%. One may find it unsettling that half (or more to some recent account) of our own genetic material is derived from genetic parasites and viruses!
Where are they?
Diverse Genome Landscapes
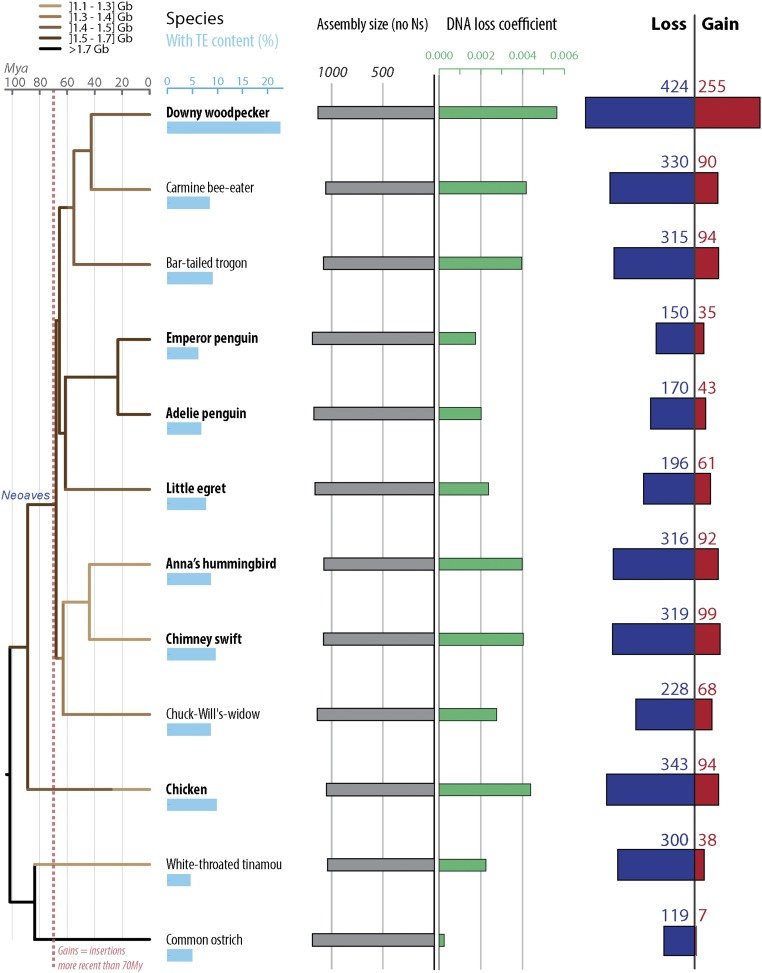
Perhaps reassuringly, most of what’s recognizable as mobile DNA sequences in the human genome are actually no longer capable of moving around. They are the remnants of previously invasive elements that have long ceased transposition activity and have now become permanent fixtures of the genome. Greater than 99.9% of human transposable elements are fixed in the population – but that still leaves plenty still jumping around and thousands that are not shared across individuals (we are keeping a close eye on those in the lab). Interestingly, this picture is very different in other species, such as mouse, fruit fly, or plants like maize. In those species and in fact probably most species (humans seem a bit exceptional that way) we see a much larger fraction of transposable elements bouncing around from one generation to the next, caught in the midst of spreading in populations. It is poorly understood why some species like humans have few active elements but zillions fossilized in the genome, while others have many active elements but relatively few debris lurking around their genomes. Some of our recent work gave some clues, but this is a question we continue to actively pursue. We are part human, part virus
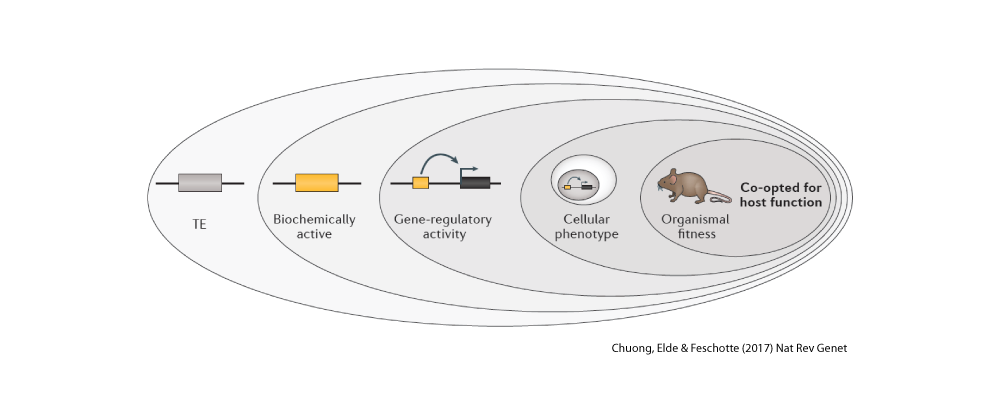
One of the most intriguing classes of mobile elements are those we can directly trace to viruses. These are often referred to as endogenous viruses because they have become part of the genetic material of the host species they once infected and as such they are now passed from parents to offspring just like the rest of the DNA. Amazingly, in our own genome we can count more genes derived from viruses than those encoding cellular proteins (i.e. proteins known to have cellular functions). However most of these viral sequences are crippled with mutations and are clearly no longer capable to encode intact viral components (in fact nobody has ever found one fully capable to encode an infectious virus – but again that story is different in other species like mice or… koala!). Endogenous viruses are one of our favorite pets in the lab. We love to play with them. I mean, we love to study them. For example, we use them as fossils to date ancient viral infections that plagued ancestral species. This emerging field of ‘paleovirology‘ provides unique insight into the mysterious origins of viruses and how they evolve on a deep timescale. We are also documenting cases where viral sequences have been repurposed for cell function for the benefits of their host species. It sounds a lot like science fiction, but it’s actually real science! Mobile DNA is often referred to as the ‘dark matter of the genome’ because we still poorly understand its functional significance. Yet the short-term, mutagenic effect of mobile elements on the structure and expression of the genome has been well documented. For example, over a hundred human diseases, such as hemophilia A and several cancers, are directly linked to de novo insertion or recombination of mobile elements. Similarly, half of all mutant phenotypes recorded in the fruit fly and more than 10% of those observed in the laboratory mouse are caused by jumping or recombining transposons. Clearly, mobile elements are potent mutagens. Much less is known about the long-term impact of mobile elements on the biology and evolution of species. This has been a major research foci of our lab for some time (see Major Question 3). In particular, we study the process by which sequences and products derived from mobile elements have been coopted as raw material during evolution to assemble new genes and regulatory elements underlying the emergence of biological innovation. Got dark matter?
OUR MAJOR QUEST(ION)S
Question 1.
What are the mechanisms allowing for the persistence of transposons? We (and many others) have uncovered flagrant cases of horizontal transfers where transposons have repeatedly crossed species boundaries to invade the genome of widely diverged animals, including mammals. But how common are these ‘jumps’ across species and how do they happen?
What is the biological significance of endogenous viruses and what do they tell us about host-virus evolution?Question 2.
Endogenous viruses can be viewed as molecular fossils that have been deposited in the genome during past viral invasions. By excavating and analyzing such genomic relics we have the opportunity to explore viral evolution at a depth unreachable when studying modern viruses. This information has the power to yield critical insights and predictions relevant to pathogenic viruses circulating now as well as those threatening to trigger the next pandemics. What is the contribution of mobile DNA to the advent of new biological functions, and in particular regulatory evolution?Question 3.
There is now compelling evidence (e.g. here) that mobile elements have profoundly influenced the evolution of genes and genomes. We are actively studying the origin and function of genes, coding and non-coding, and other regulatory sequences that have emerged from mobile elements in human, bats, zebrafish and Drosophila. We aim to assess the extent by which mobile elements have contributed to the advent of biological innovationAn Integrative Approach:
Given the ubiquitous nature of mobile DNA and the ever-growing accessibility of whole genome sequencing, we do not restrict our research to a single model species or even a group of organisms. Instead, we are studying the genomes of a range of eukaryotic species, with a current emphasis on vertebrates, including humans. Our research uses an integrative approach that lays on a foundation of bioinformatics and comparative genomics. The findings and patterns deciphered at the computer are then taken to the wet lab to test specific hypotheses in the most appropriate experimental systems, often in collaboration with other laboratories who complement our expertise (and are fun to work with!). These include functional analyses in vitro and ex vivo in mammalian cells, as well as genetic analyses in model organisms such as yeast, Arabidopsis and Drosophila. Indeed we love ALL mobile DNA!!